Elisa Gagliano: The mysteries of radical SAMs
The following story was written in May 2020 by Alyssa Jorgensen in ENGL 4824: Science Writing as part of a collaboration between the English department and the Center for Communicating Science.
Proteins are essential for life. Yet for many proteins, we don’t know exactly what they do or what function they serve. Elisa Gagliano is intent on solving the mysteries behind one superfamily of proteins: radical SAM enzymes.
Gagliano is a graduate student studying life sciences in the Department of Biochemistry at Virginia Tech. While studying biology at the University of New Mexico, Gagliano wanted to get more involved in labs and research. This eventually led her to come to Virginia Tech, as she got a research fellowship to work in David Haaks’s lab studying gene expression in wild tomatoes. She decided to return to Virginia Tech for her master’s degree, which led her to her current research journey.
Gagliano’s desire to learn more about the Archaea, bacteria that exist in conditions very similar to what we believe the earliest forms of life lived in, led her to join Anne Brown’s and Kylie Allen’s collaborative lab studying radical SAM enzymes.
What are radical SAM enzymes? Let’s begin with enzymes. Enzymes are proteins that help our bodies perform chemical reactions faster. Inspired by the educational films series Schoolhouse Rock, Gagliano describes our bodies as machines. Enzymes, she explains, are like “little machines that make chemical reactions happen more efficiently.”
“If you throw some tinker toys into the air, you can hope that they stick together,” she says to explain life without enzymes. “Or you can have a little machine that always attaches a little spool to a green rod.”
In other words, enzymes are absolutely essential for our bodies to function.
The specific function of all radical SAM enzymes is to break apart the SAM (abbreviation for S-adenosyl-L-methionine) molecule, which goes on to perform other reactions depending on the subfamily of radical SAM enzymes. Radical SAM enzymes perform this task by using a box-like structure of iron and sulfur to catch an electron and then sending the electron to break the SAM molecule.
“Without that iron-sulfur cluster, radical SAMs can’t function,” Gagliano explains.
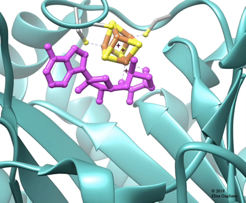
And that’s where the issue of oxygen comes in. When the Archaea first evolved, back when life began, Earth did not have an oxygen-rich atmosphere. Oxygen, it turns out, prevents the box-like structure of iron and sulfur from functioning.
As Gagliano explains, “Oxygen breaks the cluster apart—pushes out an iron molecule—which wasn’t a problem back when life began.”
Archaea today typically have a lot of radical SAM enzymes and tend to live in environments that lack oxygen, and most research on radical SAM enzymes is done on Archaea. However, many, if not most, oxygen-loving eukaryotes, including humans, also have lots of radical SAM enzymes, and we don’t exactly know why.
One important part of Gagliano’s research is comparing radical SAM enzyme counts in organisms that use oxygen and organisms that do not. Another part of her research is predicting the functions of some radical SAM enzymes based on their similarities to the radical SAM enzymes whose functions we do know.
Gagliano’s research is entirely computational. She conducts research on her computer rather than in a “wet lab,” which is what typically comes to mind when we think of lab work. Her research will eventually be used in Dr. Allen’s wet lab to test radical SAM enzymes based on Gagliano’s computational findings.
Gagliano is studying a protein found in Entamoeba histolytica, a parasitic amoeba. The purpose of this protein is unknown, but Gagliano has identified it as a potential radical SAM enzyme from a specific group, the miaB family.
Reading the scientific literature on radical SAM enzymes provides a guide to Gagliano’s research. However, most of her work involves going through public databases, such as UniProt and Pfam, and picking out radical SAM enzymes from thousands of protein sequences.
Manually, this would be a tedious task, but Gagliano makes the process easier and more efficient by using code. Code allows Gagliano to automatically pick out the radical SAM enzymes by telling her script to only pick out a certain pattern of amino acids: three cysteines (an amino acid) connected to that iron-sulfur cluster.
Gagliano’s research requires a lot of trial and error. Sometimes codes don’t work and she has to spend hours troubleshooting her scripts. Sometimes she’ll come across what seems to be a radical SAM enzyme but turns out to be a ferredoxin, a non-enzyme protein that has three cysteines but lacks some other radical SAM characteristics.
Because Gagliano’s work is predominantly computational, her research has not been significantly affected by COVID-19. She is able to do pretty much everything she needs to from her own computer.
“There are laboratory computers, but I’ve always just used my own laptop,” she explains. Gagliano is able to adapt to conducting her research completely off campus by accessing programs like Virginia Tech’s ARC computing system remotely.
Gagliano says that her research has really helped her develop her coding skills. She plans to apply those skills soon when she begins looking for jobs performing lab work and/or data analysis in fields like forensics. She’s interested in a career in which she can use her knowledge of life sciences to benefit the general public.
Gagliano has found community in the Biochemistry Graduate Student Association, enjoyed Virginia Tech’s library workshops, and done an internship doing lab work in a Coca-Cola bottling plant.
“Getting involved in research as an undergraduate, which took some persistence,” she reflected, “opened up a lot of opportunities for me.”


